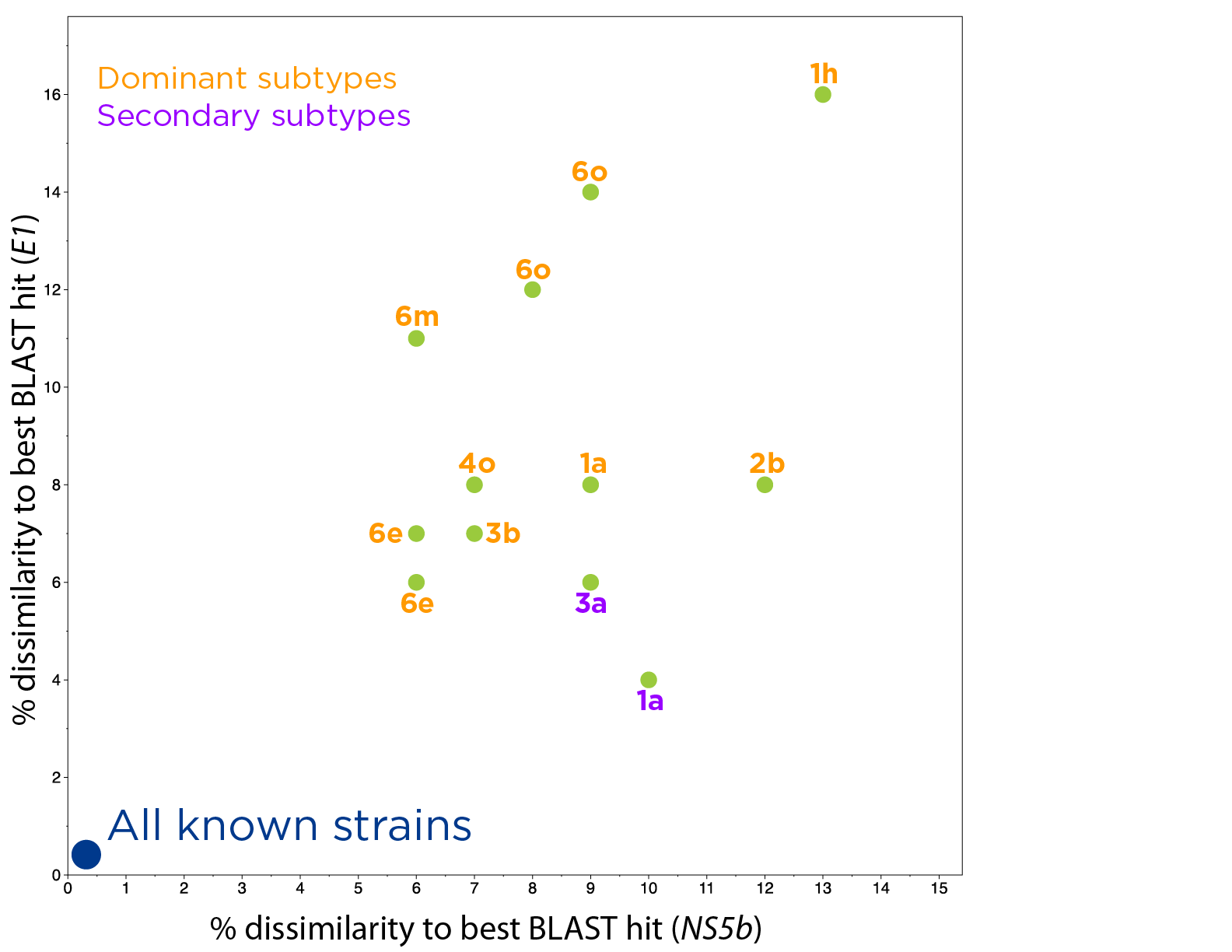
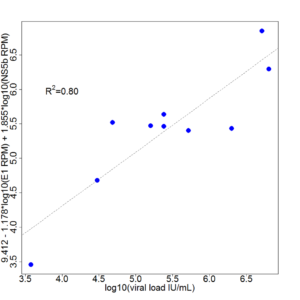
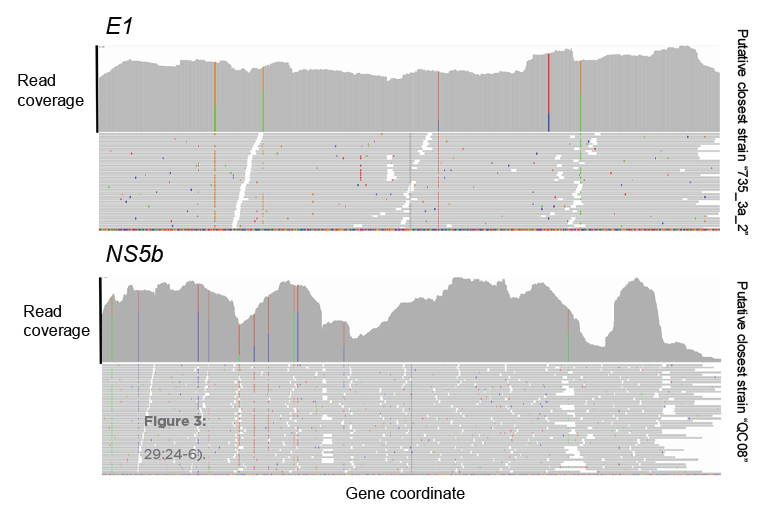
FUSION’s ONETest™ HCV assay allows for sensitive and scalable identification of Hepatitis C viruses, even in samples that fail current frontline testing.
The assay provides more genetic information than all widely employed conventional tests combined (i.e., RT-PCR assays and Sanger sequencing), including estimates of viral load and quasispecies. The assay also enables various types of epidemiological investigation—from routine in a single simple-to-perform assay.